It is of common knowledge that humans and monkeys share great similarities regarding their genomes. And, if most monkeys have tails, what happend to ours? This is the central question Xia and colleagues (Xia et al. 2024) try to answer.
After comparing the genomic sequences across primates, the researchers found a mobile genetic element called Alu only inserted on the gene TBXT of apes, a gene that encodes for a transcription factor associated with tail development in mammals. Alu elements have the ability of “teleporting” and fixating in other genome regions and therefore they are very important to diversify genome sequences. The Alu element insertion found in apes leads to an alternative splicing event, resulting in the production of a shortened form of the TBXT gene. Humans express both the full-length and the shortened versions of TBXT gene.
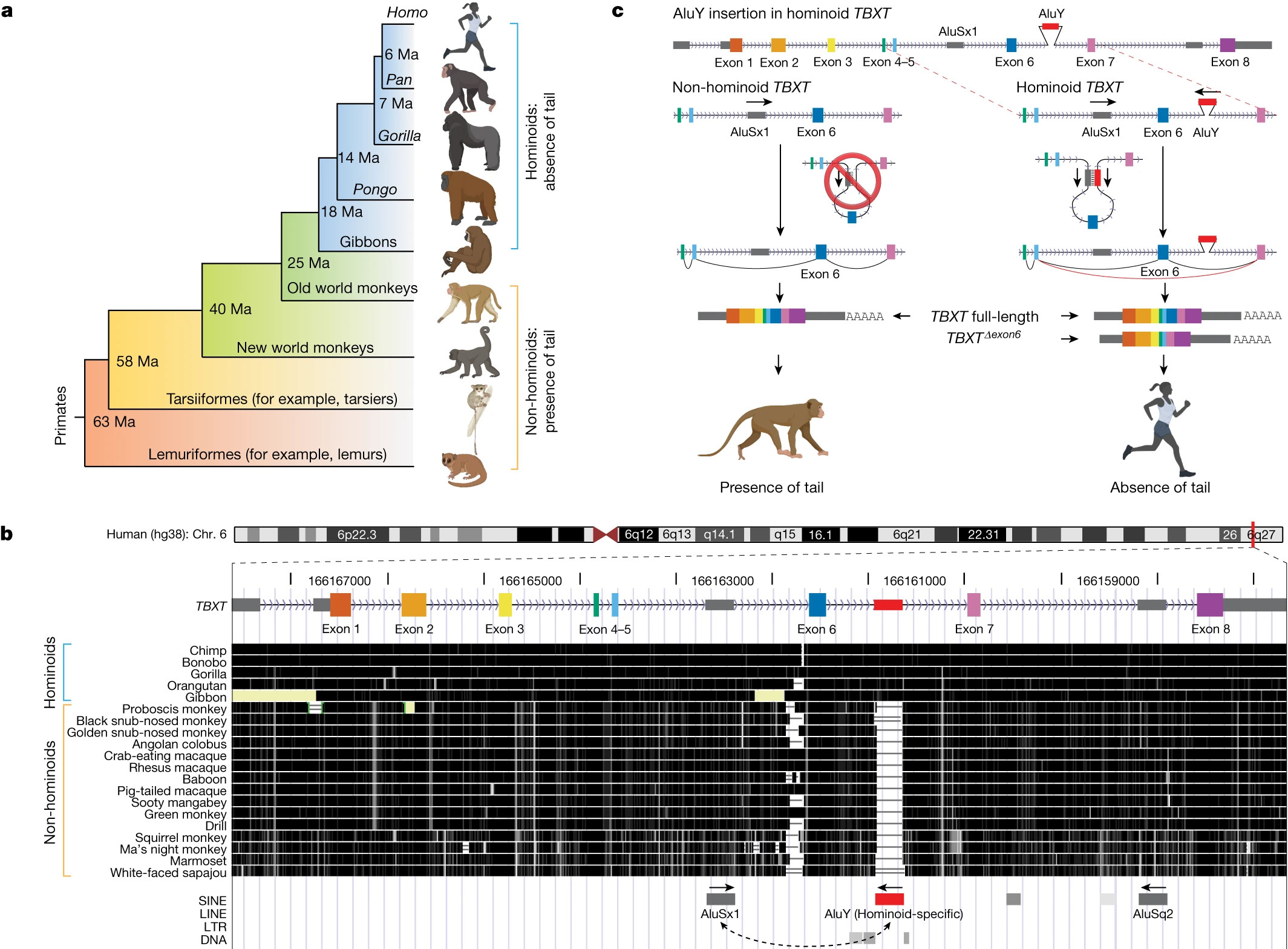
To test their hypothesis, researchers engineered mice to express these two gene versions and they observed that, in this scenario, mice exhibit a lack of a tail or a shortened tail, similar to the tail-loss phenotype in hominoids. However, for mice engineered to have only the shortened version of TBXT gene died during the embryonic development. This suggests that the evolution of tail loss in hominoids may have been associated with an adaptive trade-off, where the loss of the tail conferred evolutionary advantages but also increased the risk of neural tube defects.
This research is a great example of comparative genome analysis, common in the Bioinformatics area. Do you need help in comparing genomes? Talk to the Lexanomics team!