Severe pneumonia presents a significant challenge for cancer patients, often complicating timely and accurate diagnosis of underlying pathogens. Conventional diagnostic approaches can be sluggish and may overlook crucial infections, particularly in individuals with compromised immune systems. However, the advent of metagenomic next-generation sequencing (mNGS) is revolutionizing diagnostic protocols by expediting accurate identification and bypassing the need for additional clinical assessments such as Gram staining, serum immunological tests, G tests, GM tests, and PCR tests.
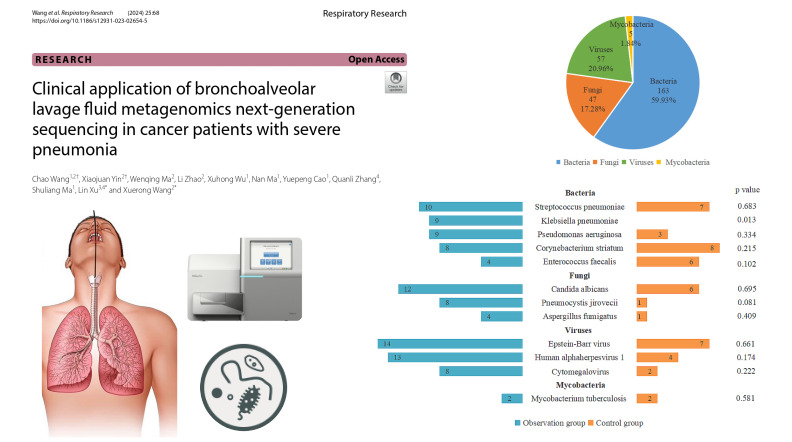
In a study targeting cancer patients grappling with severe pneumonia, Wang et al. (Wang et al. 2024) harnessed mNGS to scrutinize lung fluid in conjunction with standard culture techniques. Their findings underscored mNGS’s superior efficacy in detecting infections, even discerning multiple microorganisms simultaneously. Notably, it excelled in identifying elusive pathogens like Streptococcus pneumoniae and Aspergillus, which often elude traditional diagnostic methods.
The research demonstrated that mNGS results prompted treatment adjustments for over half of the patients, leading to improved clinical outcomes. By furnishing clinicians with precise information regarding the causative agents, mNGS facilitated targeted antibiotic or antifungal therapy selection.
Although mNGS shows great promise, it does have some drawbacks. It can be expensive, and there are still technical issues to work out. Also, because it’s very sensitive, it can’t always tell if something is contamination, colonization, or an actual infection. Still, the study shows that mNGS can really help doctors diagnose and treat infections in cancer patients with severe pneumonia. This new tool helps healthcare workers handle these serious illnesses better, which should lead to better outcomes in the future.