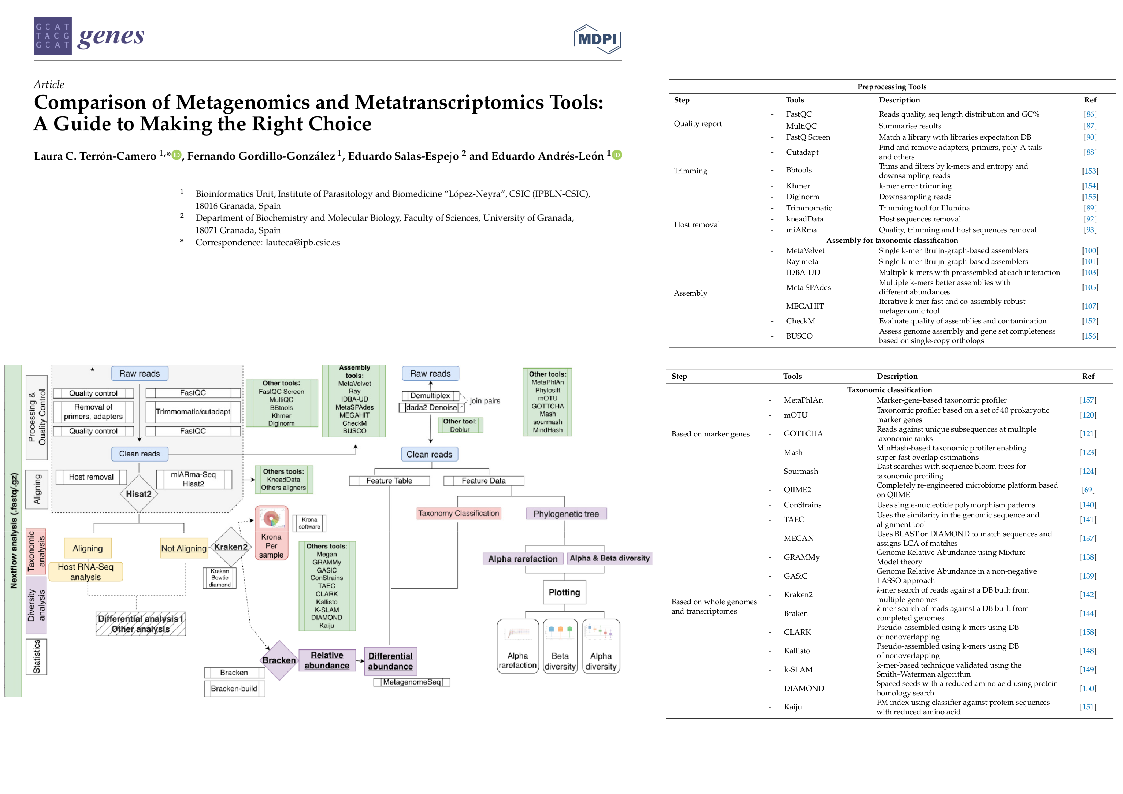
Bioinformatics is an area where new analytical methods, tools, and pipelines are constantly emerging. Therefore, knowing which tools are most appropriate for your data is essential to ensure the high quality of analyses and reproducibility.
With the aim of providing an overview of the techniques and tools available for Metagenomics and Metatranscriptomics, both from a methodological and bioinformatics perspective, Terrón-Camero and collaborators (Terrón-Camero et al. 2022) conducted a series of comparisons between read classification tools such as QIIME2, MEGAN, mOTU, MetaPhAn, Kranken2, and others; and also provides pipelines to execute all of them in an automated manner, using Nextflow.
The authors provide some tips on how to proceed with read classification, and here is a short answer:
- For shotgun metagenomics: Kraken2;
- For 16S analysis: we recommend using the QIIME 2 tool when a low number of reads exists;
- Also for 16S analysis: Kraken2 is efficient when the library was deeply sequenced.
- For metatranscriptomics data: Kraken2 and Bracken.
The work also makes it very clear the need to perform filtering by applying a threshold to eliminate organisms with low abundance, thus reducing false positives.
In addition to the technical characteristics of the tools, the article also provides a comprehensive review of the influence of gut microbiota on various conditions that can affect humans, which further enhances the value of reading the paper.