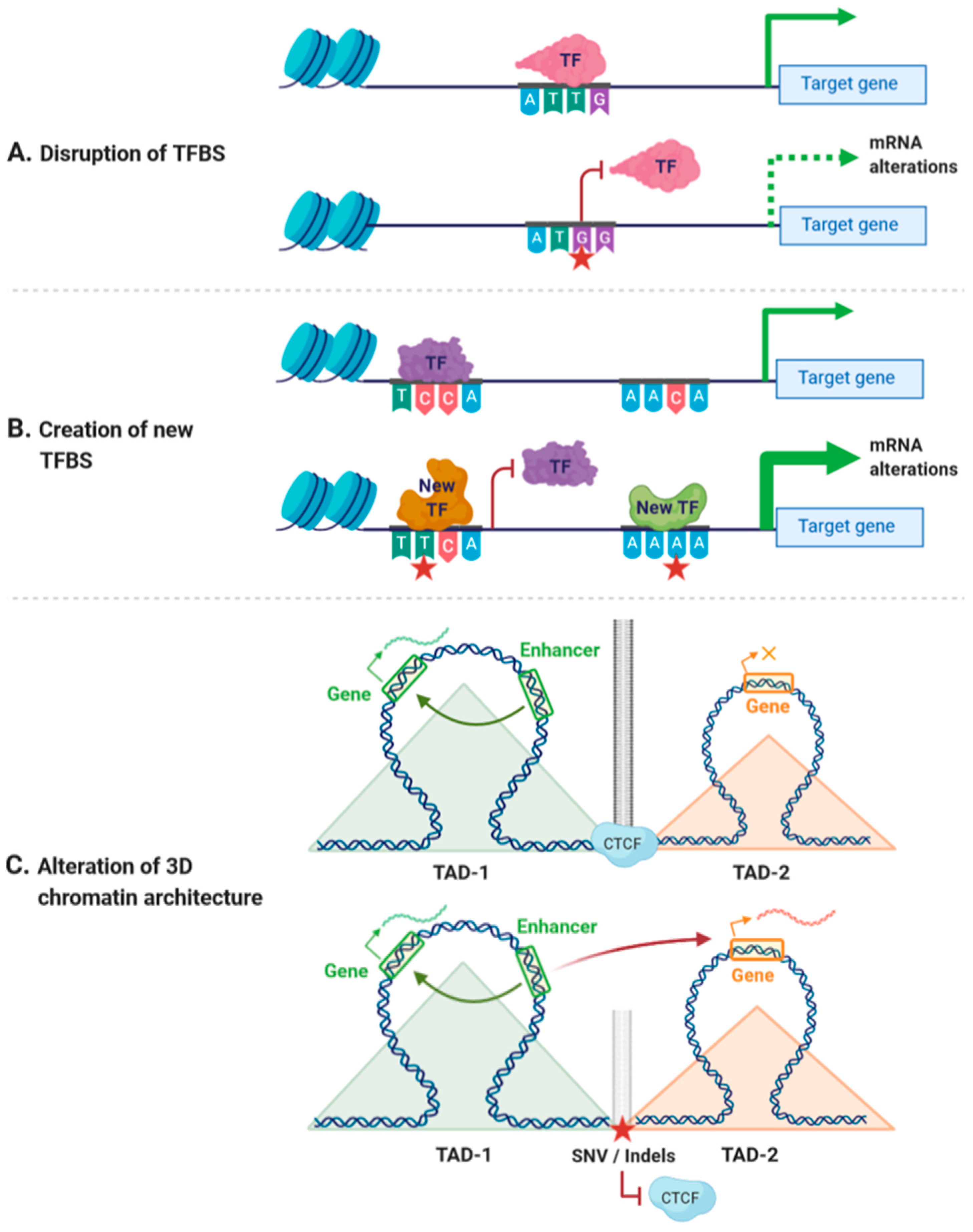
Coding variants, especially single-nucleotide variants, are genetic alterations occurring within the coding region of the genes. They are widely studied in human genomics given their relationship to human diseases, including rare Mendelian disorders and complex diseases. The effects of coding variants that result in a change of amino acids (e.g., missense and nonsense variants) can be promptly identified in the protein sequence, impacting the protein function. However, what are the effects of non-coding variants in human diseases?
Developmental disorders are mental and/or physical disabilities typically starting in infancy. About 30-40% of individuals with DD are associated with a genetic diagnosis, indicating that the condition has also a multifactor background. In a new pre-print, Lord et al. analyzed the data of probands and relatives from Genomics England 100,000 Genomes and investigated the genetic architecture of DD (Lord et al. 2023). In the study, the researchers focused on identifying rare non-coding variants in trans (inherited from the alternate parent to the coding variant) that could potentially act as the “second hit” in compound heterozygous genotypes associated with developmental disorders. They defined and analyzed various non-coding regulatory regions associated with the 793 identified recessive genes, including intronic regions, 5’UTRs, 3’UTRs, and candidate upstream promoter regions.
After stringent bioinformatic filtering and quality control measures, the researchers identified rare non-coding variants in these regulatory regions that passed their criteria. These non-coding variants were assessed for their potential regulatory impact on gene expression and function. The study found that a subset of proband-variant pairs (36.3%) had at least one rare non-coding variant in trans that met the quality filters and could potentially contribute to the pathogenicity of the genotype.
Furthermore, the researchers conducted functional testing using RNA sequencing to evaluate the impact of these non-coding variants on gene expression and splicing. This approach provided insights into how the identified non-coding variants may disrupt regulatory elements and contribute to the pathogenesis of developmental disorders in conjunction with coding variants in recessive genes. In addition, this study sheds light on the potential role of non-coding variants on the physiopathology of other conditions.