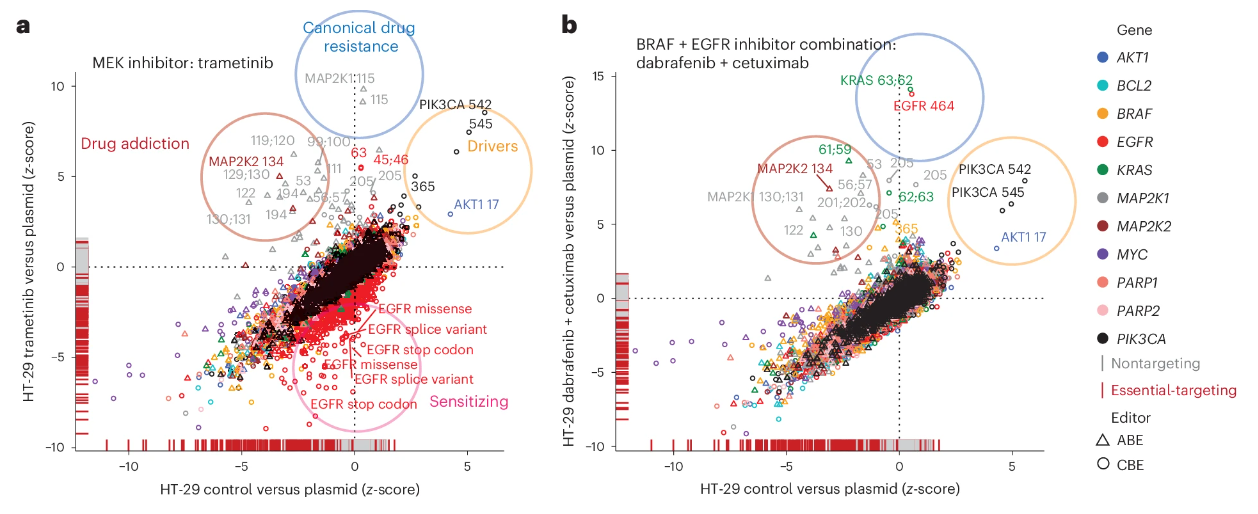
Recent advancements in cancer research have focused on understanding the genetic mechanisms that contribute to drug resistance in cancer therapies. Using CRISPR base editing mutagenesis screens, researchers have identified,476 variants across cancer genes in four different cancer cell lines. This work has resulted in the classification of four functional classes of protein variants that modulate drug sensitivity. These classes include ‘drug addiction variants,’ which provide a proliferation advantage in the presence of drugs but are detrimental without them, and ‘canonical drug resistance variants,’ which confer direct resistance to specific therapies.
The study underscores the importance of systematically annotating variants to enhance the understanding of drug resistance mechanisms. High-throughput approaches allow for the rapid functional evaluation of variants, particularly those of unknown significance. By employing base editing, researchers can create a comprehensive map detailing how specific variants affect drug sensitivity and resistance, thereby informing clinical management strategies for patients with drug-resistant cancers. This methodology not only accelerates the identification of drug resistance mechanisms but also aids in the development of new inhibitors targeting resistant proteins.
Furthermore, the research highlights the integration of single-cell transcriptomics to elucidate the distinct mechanisms by which these variants operate. For example, the effects of variants can be observed through differential gene expression analysis, enabling the identification of pathways involved in drug resistance. This approach could lead to more effective therapeutic strategies and improved patient outcomes by tailoring treatments based on the specific genetic profiles of cancers.