Generative AI has garnered significant attention in bioinformatics. Techniques like Generative Adversarial Networks (GANs) and Variational Autoencoders (VAEs) are at the forefront, reshaping how we analyze complex biological data.
Today, let’s delve into Variational Autoencoders (VAEs) and their groundbreaking application in identifying drug–omics associations for type 2 diabetes.
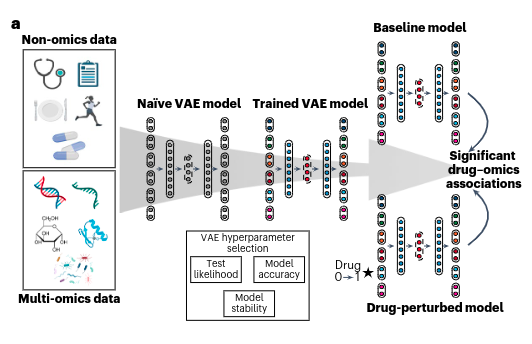
If you’re unfamiliar with the concept, VAEs are a type of neural network designed to simplify complex data. They transform high-dimensional datasets—such as multi-omics data—into a streamlined, more manageable “latent space.” This latent space captures essential patterns while filtering out noise, making it easier to uncover meaningful relationships.
Now that we’ve covered the basics of VAEs, let’s introduce MOVE — a generative model for drug-omics association analysis.
MOVE (Multi-Omics Variational Autoencoders) is a deep-learning framework developed to tackle a critical challenge in bioinformatics: integrating multi-omics data.
In their study, Allesøe et al. utilized a comprehensive dataset encompassing genomics, transcriptomics, proteomics, metabolomics, microbiome profiles, and even clinical and lifestyle information.
What makes MOVE stand out? It’s not just about analyzing data—it’s about making predictions and discovering hidden connections. By leveraging its generative capabilities, MOVE can simulate how drugs might affect patients who have not yet received them. Think of it as a virtual laboratory experiment, uncovering potential drug–omics interactions that would otherwise remain concealed in the complexity of the data.
MOVE was applied to data from 789 newly diagnosed type 2 diabetes (T2D) patients, yielding transformative insights:
- Revealing how metformin (a common diabetes medication) influences the gut microbiota, offering new perspectives on its effects beyond glycemic control.
- Highlighting distinct molecular responses to statins (cholesterol-lowering drugs), specifically simvastatin and atorvastatin, which are often considered interchangeable.
- Detecting over 200% more drug–omics associations compared to traditional statistical methods.
Are you exploring Generative AI in your research? If so, we’d love to hear about your experiences!